
index/Resource
Tools
metaMIC
期刊机构:Genome Biology
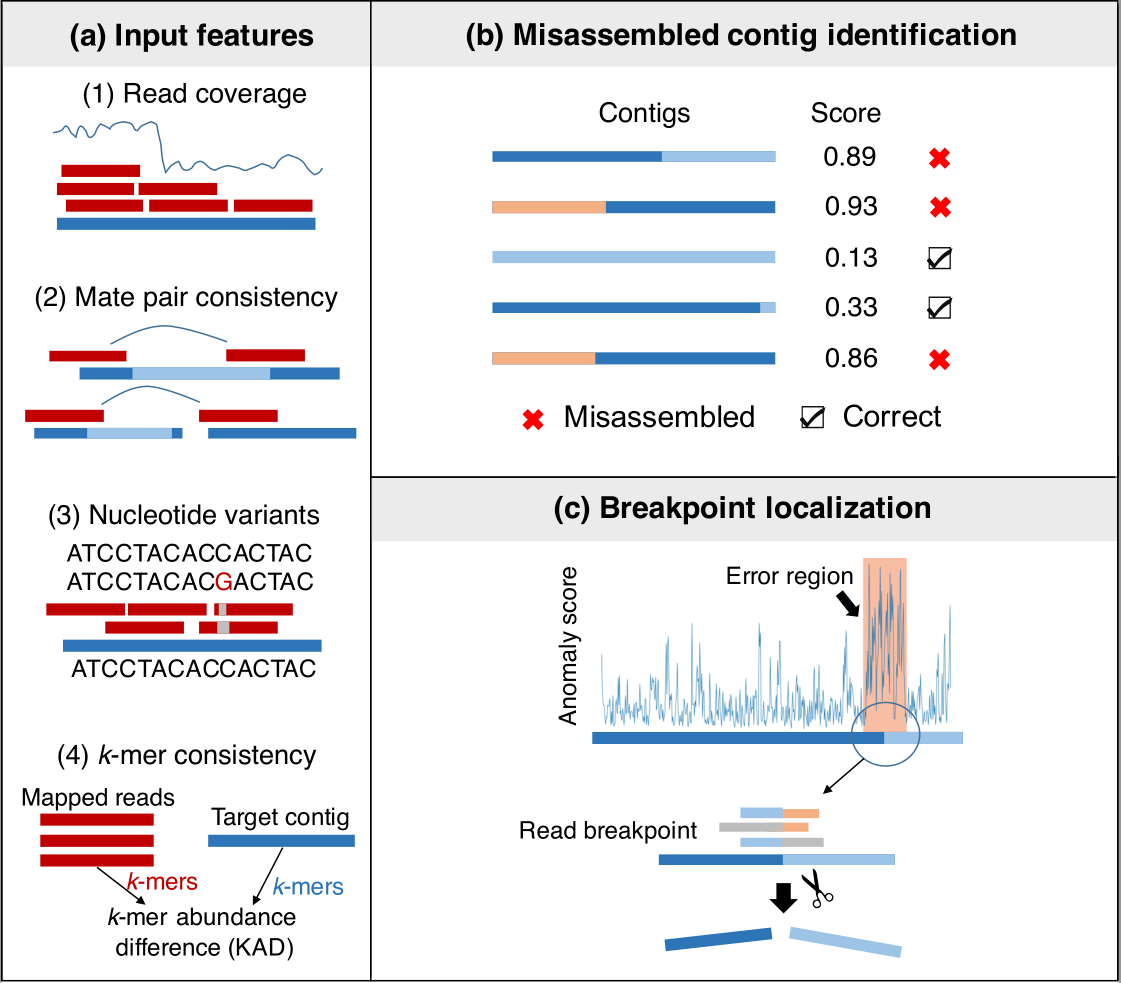
Firstly, metaMIC extracts various types of features from the alignment between paired-end sequencing reads and the assembled contigs.
Secondly, the features extracted in the first step will be used as input of a random forest classifier for identifying misassembled metagenomic assemblies. Thirdly, metaMIC will localize misassembly breakpoints for each misassembled contig and then correct misassemblies by splitting into parts at the breakpoints.
Source Code:https://mai.fudan.edu.cn/webimgs/toos/metaMIC/metaMIC-main.zip


