
工具
nMAGMA
期刊机构:Briefings in Bioinformatics
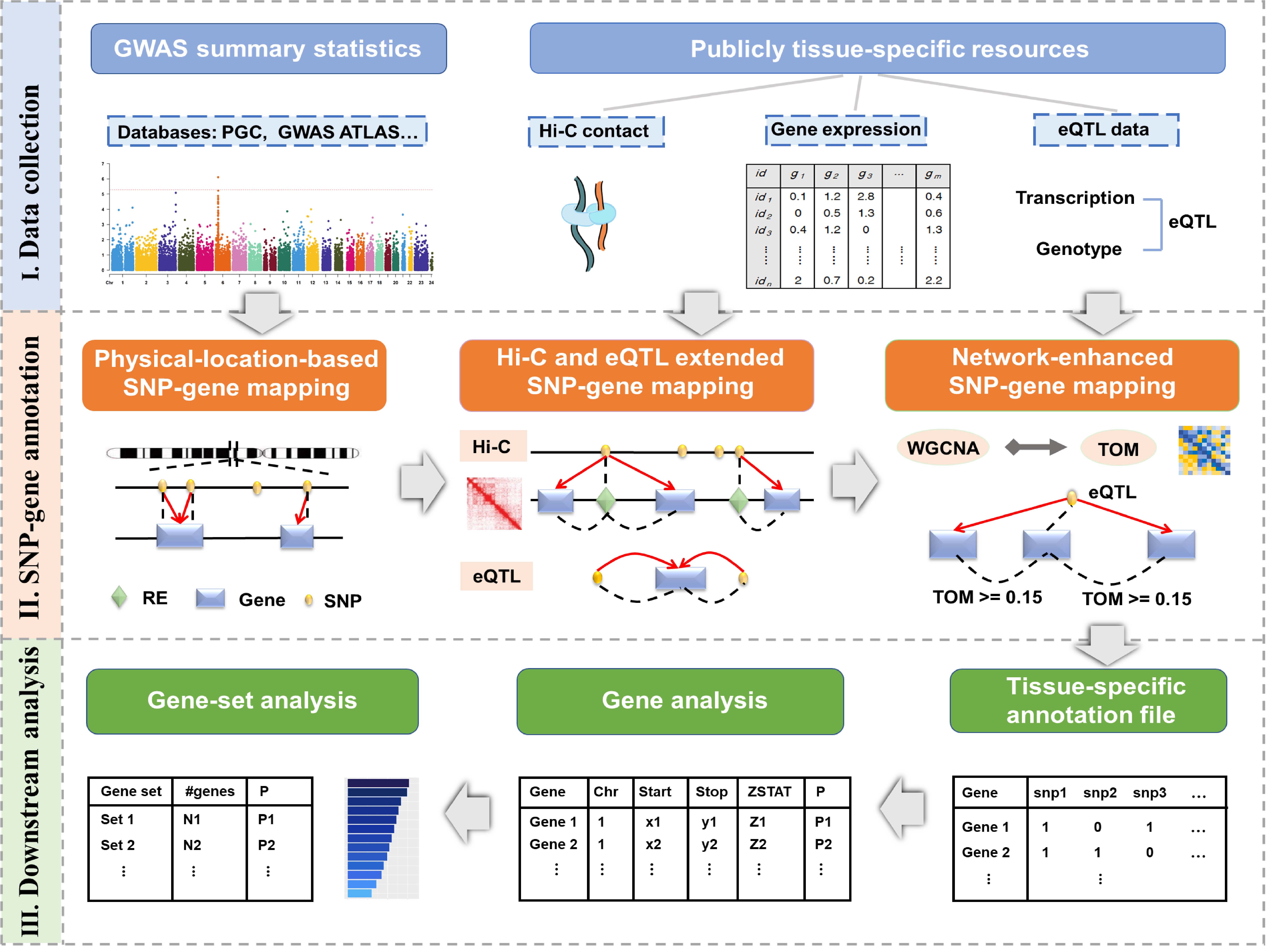
In nMAGMA, we inferred risk genes from SNPs with the following steps: Firstly, the SNPs were mapped to genes and regulatory elements (REs) using MAGMA based on their physical locations, where a 0kb-window was adopted for the European population; Secondly, the SNPs were assigned to genes via SNP-to-RE annotations and RE-gene regulatory pairs derived from Hi-C-based chromatin interaction profiles; Thirdly, the SNPs were assigned to genes based on significant eQTL results from GTEx; Finally, the topological overlap matrix was calculated with WGCNA for genes based on their expression profiles from GTEx, where a pair of genes with a TOM ³ 0.15 were regarded as strongly interconnected as described previously. In this way, the lists of genes inferred from the first three steps can be further extended, where a gene that has a TOM ³ 0.15 with any gene inferred from the first three steps will be included in analysis. Finally, we got a final SNP-gene mapping annotation file, with which we can perform gene analysis to determine the association of a certain gene with a disease, as well as perform gene-set analysis to determine the association of a certain gene set with a disease.
Source Code: https://mai.fudan.edu.cn/webimgs/toos/nMAGMA/nMAGMA-master.zip


